Lava Caves of Hawaiʻi
Deep inside the slopes of Kīlauea, unique communities of extremophilic bacteria thrive within the inhospitable crevices of caves formed by once flowing lava. This unseen and often uncharacterized biodiversity may hold new compounds of antibiotic or pharmaceutical interest, or reveal ways in which early life on Earth found purchase in a primordial world.
Banner image credit: Kenneth Ingram
Biofilms inside a Hawaiʻi Island lava cave.
Photo credit: Jimmy Saw
Moanalua High School students prepare to sequence a genome using the ʻĀina-Informatics mobile lab.
Photo credit: ʻIolani School
Across Hawaiʻi Island, lava caves have also served as important cultural landscapes that throughout history have offered shelter, fresh water, refuge from conflict and sacred protection for ʻiwi kupuna. A dazzling array of microbes inhabit these caves, some able to survive off just the lava rock itself, converting the very pele into the ʻāina momona which sustains us all.
Wāhine preparing wauke and pounding kapa at a cave entrance.
These unique microbes have been painstakingly collected and cultured from these caves under the direction of microbiologists Dr. Stuart Donachie (University of Hawaiʻi at Mānoa) and Dr. Rebecca Prescott (NASA Postdoctoral Fellow) for decades.
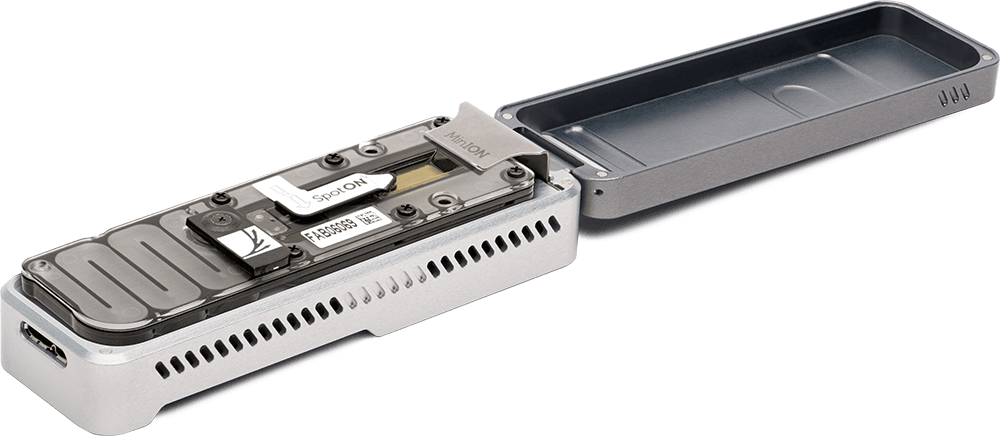
Since 2018, students within the ʻĀina-Informatics Network have used cutting-edge MinION sequencing technology and bioinformatics tools to assemble and explore the genomes of these rare cultures.
Novel Species
For some of these bacteria, students discover that the species they have sequenced has not previously been described to Western science. In these cases, the ʻĀina-Informatics network has been generously offered the opportunity by our UH partners to propose scientific names for consideration by the International Committee on Systematics of Prokaryotes (ICSP).
Hawaiʻi Preparatory Academy students training on the Oxford Nanopore MinION sequencer.
Our Academic Partners
This project is a collaboration between ʻĀina-Informatics, the Donachie Lab in the School of Life Sciences at the University of Hawaiʻi at Mānoa, Los Alamos National Laboratory, the NASA Exobiology Program and NASA Johnson Space Center. Mahalo nunui to the many researchers past and present who have contributed to the discovery and cultivation of the bacteria sequenced in this effort.
Dr. Stuart Donachie
is a Professor of Microbiology at the University of Hawaiʻi at Mānoa. Dr. Donachie specializes in the collection, cultivation and characterization of microbial diversity and function from diverse environments throughout the Hawaiian Islands and beyond.
Dr. Rebecca Prescott
is currently a NASA Postdoctoral Fellow working with the NASA Johnson Space Center and her home institution, the University of Hawaiʻi at Mānoa. As a resident of the Hawaiian Islands for the past 15 years, Dr. Prescott’s research focuses on microbial interactions in environments from volcanic islands, and human cultural connections to those microbial communities.
Lava Cave Microbes Lab
Students use the MinION to sequence cultured microbial isolates from Hawaiʻi Island lava caves. This lab is designed to sequence three targets simultaneously on a standard MinION flow cell, but it can be modified to do a single genome or a duplex run. Flongle protocol coming soon.
LEARNING OBJECTIVES:
Students learn first, second and third generation sequencing techniques to connect genetics to genomics
Assemble a genome using basic online bioinformatics tools
Investigate what genomic data can reveal about an organism’s function or capabilities
Crosscutting concepts: DNA replication, gene expression, computer algorithms, taxonomy
Lab techniques: MinION sequencing, online bioinformatics
Keywords: Microbial biodiversity, early Earth, astrobiology, drug discovery, extremophile biology, quorum sensing
HANDOUT: MinION SEQUENCING LAB ROADMAP and TEACHER GUIDE
ACTIVITY: GENOME ASSEMBLY PUZZLE
UNIT 1: MinION SEQUENCING
Library prep protocol | MinION protocol
UNIT 2: WHOLE GENOME BIOINFORMATICS
Bioinformatics
In order to determine if these lava cave bacteria are new species and to understand if they have special genes that allow them to thrive in these extreme environment we first need to “assemble“ the bacterial genome from the DNA we sequence. Imagine the genome as a puzzle; the DNA sequences are the pieces we need to put them together to complete the puzzle.
While this sounds like a daunting task, our aim was to make this process as simple as possible, without the hassle of worrying about software version compatibility and the ever-persistent headache of installing new software. As part of our mission, we aim to make bioinformatics as user friendly as possible with the emphasis that we all have the ability to conduct this type of analyses.
Introducing the Unicellular Long-read Assembly aNd Annotation (Ulana) bioinformatics pipeline. Ulana was developed in-house to provide a fast, convenient, and user-friendly option for assembling and annotating bacterial genomes. The Ulana pipeline can run on any Windows, Mac or Linux based device and is distributed as a docker image; as long as you have docker installed, you can execute the pipeline from any computer. Unfortunately, at this time, Ulana is not compatible with chromebooks or ChromeOS based devices. However, a Ulana server can now be accessed directly using any device via a web browser, furthering reducing teachers’ barriers to access.
Publications
Bridging place-based astrobiology education with genomics, including descriptions of three novel bacterial species isolated from Mars analog sites of cultural relevance
Rebecca D. Prescott, Yvonne L. Chan, Eric J. Tong, Fiona Bunn, Chiyoko T. Onouye, Christy Handel, Chien-Chi Lo, Karen Davenport, Shannon Johnson, Mark Flynn, Jennifer A. Saito, Herb Lee Jr., Kaleomanuʻiwa Wong, Brittany N. Lawson, Kayla Hiura, Kailey Sager, Mia Sadones, Ethan C. Hill, Derek Esibill, Charles S. Cockell, Rosa Santomartino, Patrick S.G. Chain, Alan W. Decho, and Stuart P. Donachie
Astrobiology. 2023. 23(12): doi: https://doi.org/10.1089/ast.2023.0072